Creating Easy-To-Use Bioinformatics Templates
May 2025 – August 2025
Principal Investigators: Dr. Arun Seetharam, Dr. Nannan Shan
Institution: Purdue University
Department: Rosen Center for Advanced Computing
Research Focus
This research project focused on making bioinformatics workflows more accessible for biologists who need to use High-Performance Computing (HPC) for large-scale data analysis.
Existing workflow management tools—such as Nextflow and Snakemake—are powerful but require learning a Domain-Specific Language (DSL), which creates a steep learning curve for many users.
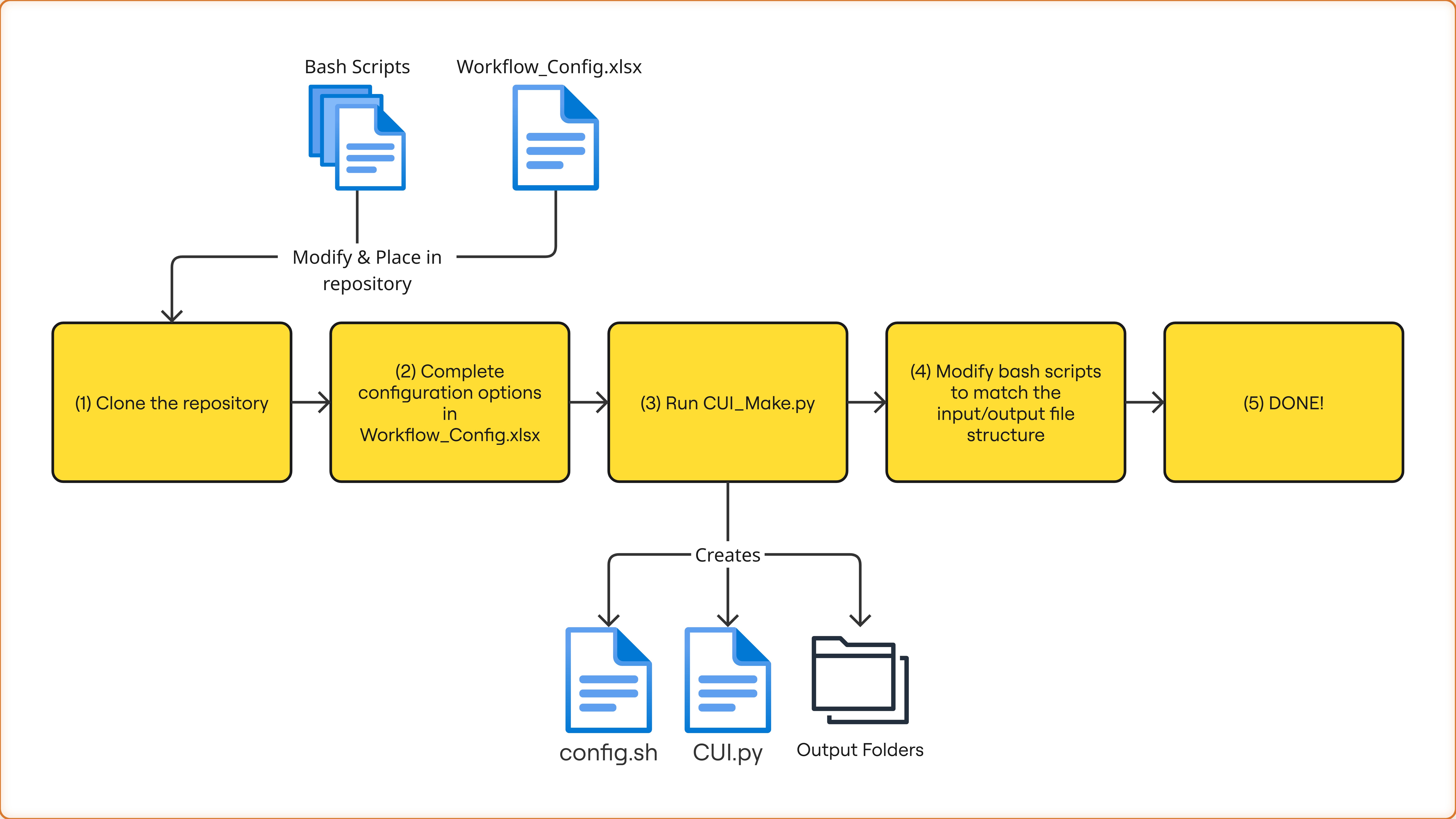
Our approach reduced this barrier by creating workflow templates that rely only on Python and Bash, two languages most researchers already know.
These templates enabled users to perform complex bioinformatics analyses without needing to master specialized frameworks.
The workflows developed included:
- Hifiasm and Flye for genome assembly
- GATK for variant calling
- RNA-seq analysis pipelines
These tools provided flexible, ready-to-run workflows designed to simplify HPC use in biological research.
Responsibilities
I worked with Dr. Arun Seetharam and Dr. Nannan Shan at Purdue University’s Rosen Center for Advanced Computing (RCAC) as part of a summer REU internship, under the supervision of Arman Pazouki, the project’s overall PI and NSF grant holder.
My primary responsibilities included:
- Programming and developing workflow templates for Hifiasm genome assembly and GATK variant calling using Bash and Python
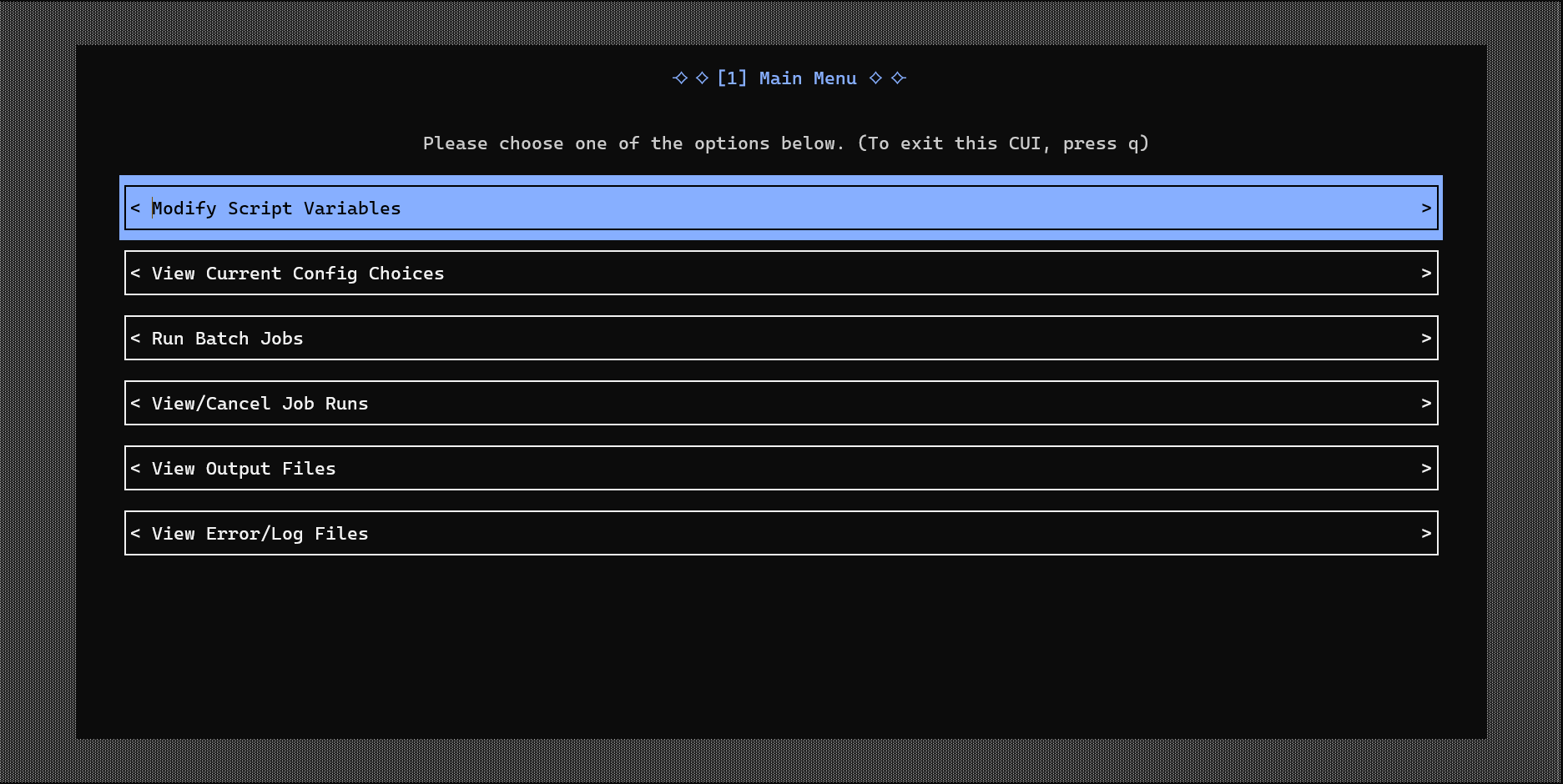
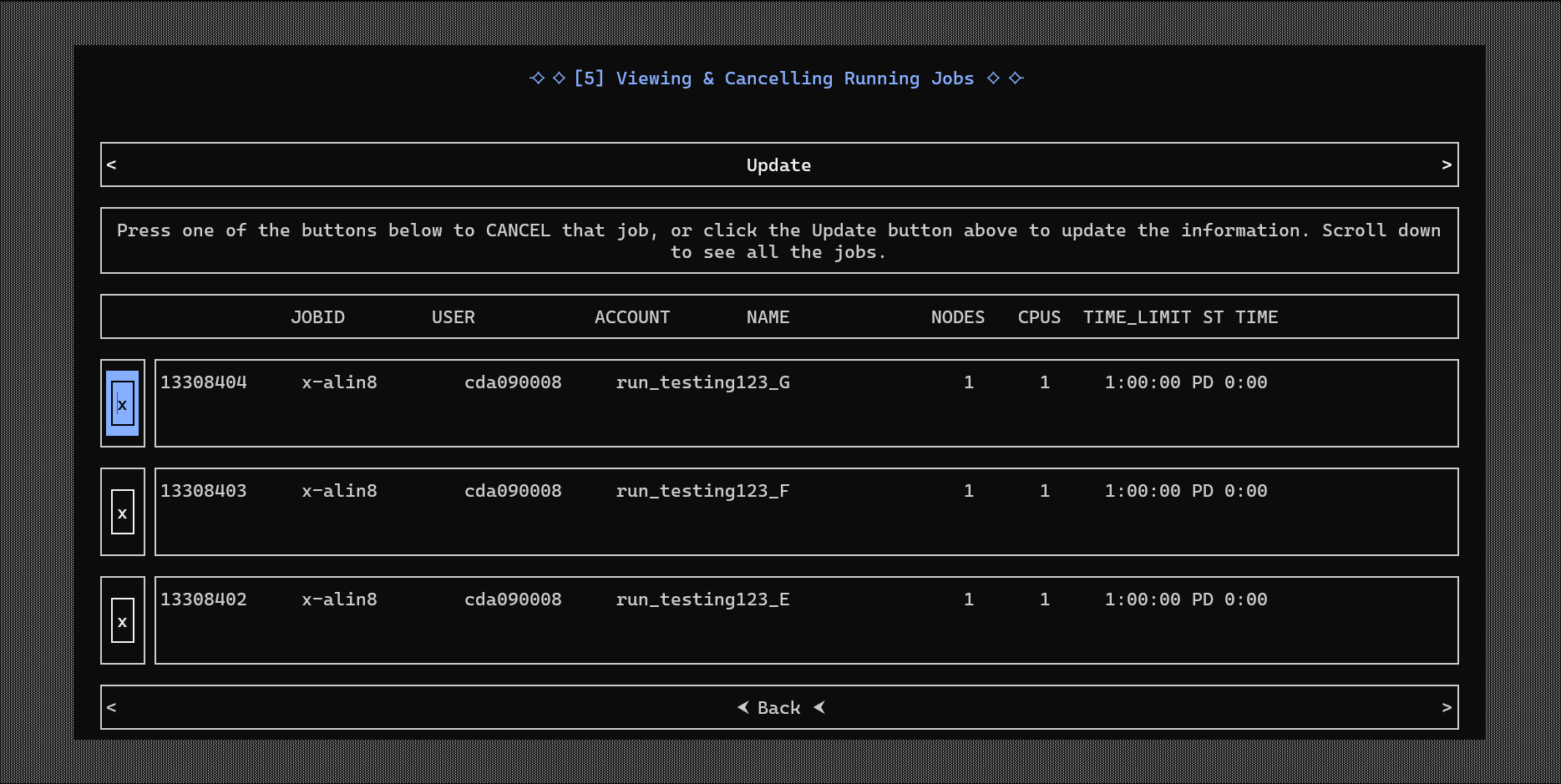
- Building a Console User Interface (CUI) with Urwid, allowing users to interactively manage workflows
- Designing a generalized template generator that automatically created new CUIs from user-defined pipelines
These contributions helped produce modular, user-friendly tools that broaden access to high-performance bioinformatics computing.
Example Workflow Visuals
By me
Workflow Diagram